Solutions Q53 - 61
Contents
Solutions Q53 - 61#
# import all python add-ons etc that will be needed later on
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
init_printing() # allows printing of SymPy results in typeset maths format
plt.rcParams.update({'font.size': 16}) # set font size for plots
Q53 answer#
Normalizing \(\vec n=\vec b\times\vec c=|\vec b||\vec c|\sin(\alpha)\vec u_n\) gives \(\vec n\cdot\vec n = |\vec b |^2|\vec c |^2\sin^2(\alpha)\vec u_n\cdot\vec u_n\).
The dot product of two equal unit vectors is \(\vec u_n\cdot\vec u_n =1\), therefore equation \(\vec u_n\cdot\vec u_n=|\vec b|^2|\vec c|^2\sin^2(\alpha)\) and the unit vectors are not present. A similar equation applies for \(m\).
Q54 answer#
(a) To define the plane \(C_1-C_2-C_6\) the vectors can be chosen to be \(\vec d_1\) and \(\vec d_2\), and for the other plane \(\vec d_2\) and \(\vec d_5\), see figure 46. The fact that \(\vec d_5\) does not join the end of \(\vec d_2\) does not matter because it is still in the \(C_1-C_2-C_6\) plane and points in the correct direction. Any other vector in this plane would do. Calculating the vectors as coordinates means defining an origin and basis set.
The coordinates of only three atoms need to be found in this particular molecule because, by symmetry, the others follow. Starting with atom \(C_2\), this is positioned at \(x = -d/2\), because the length of any side is \(d\) and it is positioned midway along \(C_2-C_6\) in the positive y direction. The \(z\) coordinate is zero because the atom is in the \(x-y\) plane, as are \(C_3, C_5\) and \(C_6\). The length \(C_2-C_6\) is, by using the cosine rule on triangle \(C_2-C_1-C_6\), \(\sqrt{2d^2-2d^2\cos(\theta)}=d\sqrt{8/3}\) because \(\cos(\theta)=-1/3\). The atom \(C_2\) therefore, has half this length as its \(y\) coordinates and \(C_2 = (-d/2,d\sqrt{2/3},0)\) and by symmetry
Some vectors can now be made. Vector \(\vec d_5\) is \(C_5 - C_6\) and is \(d_5 =d\;\boldsymbol i+0;\boldsymbol j+0\;\boldsymbol k\) and \(\vec d_2 =C_2 -C_6 =0\;\boldsymbol i+2d 2/3\;\boldsymbol j+0\;\boldsymbol k\).
Next, calculate the coordinates of atom \(C_1\). Let a vector \(\vec p\) be that from \(C_6 \to C_1\) and is written as \(p=d(a\;\boldsymbol i+b\;\boldsymbol j+c\;\boldsymbol k)\) where \(a, b\) and \(c\) have to be found; this definition means that \(\vec p\) points away from \(C_6\) and that its coefficients are normalised making \(a^2 +b^2 +c^2 =1\). The dot product between \(\vec p\) and \(\vec d_5\) is
therefore \(a = -1/3\) and therefore \(b^2 + c^2 = 8/9\).
The dot product with \(\vec d_2\) will allow \(c\) to be calculated;
and as \(b = \cos(\pi/2-\theta/2)=\sqrt{2/3}\) and therefore \(c=\sqrt{2/3}\).
The y coordinate of atom \(C_1\) is zero because by symmetry it lies on the \(x\)-axis. The coordinates of \(C_1\) are found using the definition of \(\vec p\) which is \(\vec p = \vec C_1 -\vec C_6\) therefore \(\displaystyle C_1=(-5d/6,0,\sqrt{2/3}d)\).
Vector \(\vec d_1\) should point towards \(C_6\) and therefore its components are calculated from \(\vec C_6-\vec C_1\) making \(\vec d_1=(d/3)\;\boldsymbol i-\sqrt{3/2}d\;\boldsymbol j-\sqrt{2/3}d\;\boldsymbol k\) which is the inverse of \(\vec p\).
Now that all the atoms coordinates are known, or inferred by symmetry, the rest of the question can be answered.
(b) The distance between the \(C_1\) and \(C_4\) atoms is easily calculated since their coordinates are known. The coordinate of \(C_4\) in the boat form, has the same \(z\) value as \(C_1\) in the chair form; \(C_{1,boat} = ( -5d/6, 0, \sqrt{2/3}d )\) but in the chair form \(C_4 = -C_1\), therefore \(C_{4,chair}=(5d/6,0,-\sqrt{2/3}d)\), and the \(C_1 - C_4\) distance is \(7d/3=2.3^\cdot\). In the boat form, only the \(x\) and \(y\) values are changed, \(C_{4,boat} = (5d/6, 0, 2/3d)\) and the distance \(5d/3\).
d = symbols('d', positive = True) # Use sympy make d a real positive value
C1 = Matrix( [-5*d/6,0 , d*sqrt(2/3)] ) # coordinates 1-6 for chair form.
C2 = Matrix( [-d/2 , d*sqrt(2/3) , 0] )
C3 = Matrix( [d/2 , d*sqrt(2/3) , 0] )
C4 = Matrix( [5*d/6 , 0 , -d*sqrt(2/3)] )
C5 = Matrix( [d/2 , -d*sqrt(2/3), 0] )
C6 = Matrix( [-d/2 , -d*sqrt(2/3), 0] )
C4b= Matrix( [5*d/6 , 0 , d*sqrt(2/3)] ) # C4 boat
C14chair = sqrt((C1-C4).dot(C1-C4))
C14chair

C14boat = sqrt( (C1 - C4b).dot(C1 - C4b) )
C14boat
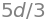
(c) The \(C_1-C_6-C_2\) and \(C_2-C_6-C_5\) dihedral angles. Consulting figure 46 and defining the normal vector \vec n = \vec d_2 \times \vec d_5$, gives
making \(|\vec n|=\sqrt{8/3}d^2\). The other perpendicular vector \(\vec m\) is
In the final dot product, only the \(\boldsymbol k\) vector term is not zero, using \(\displaystyle \cos(\psi)=\frac{\vec m\cdot\vec m}{|\vec n||\vec m|}\) we find the dihedral angle \(\psi =112.2^\text{o}\).
# using sympy with previously defined values.
d1 = C6 - C1
d2 = C2 - C6
d5 = C5 - C6
m = d1.cross(d2)
n = d2.cross(d5)
psi = acos(m.dot(n) /(sqrt(m.dot(m))*sqrt(n.dot(n)) ) )
print('{:s}{:8.2f}'.format('dihedral angle = ', psi*180/np.pi ))
dihedral angle = 112.21
(d) In calculating the torsion angle between \(C_1-C_6-C_2\) and \(C_1-C_6-C_5\), the atoms \(C_1C_6\) join the two planes involved. The vector \(\vec m\) is therefore \(\vec m=-\vec d_2 \times \vec d_1\) and \(\vec n=\vec d_1 \times \vec d_5\). The calculation is
m = -d2.cross(d1)
n = d1.cross(d5)
psi = acos(m.dot(n) /(sqrt(m.dot(m))*sqrt(n.dot(n)) ) )
print('{:s}{:8.2f}'.format('dihedral angle = ', psi*180/np.pi ))
dihedral angle = 74.50
(e) The angle between \(C_1-C_6-C_2\) and C\(_5-C_4-C_3\) in the boat form, requires us to use \(C_{4b}\) as defined above. The vector \(\vec d_4\) can be translated to start at \(d_2\) without changing the torsion angle.
d4 =C4b - C5
m = d1.cross(d2)
n = d2.cross(d4)
psi = acos(m.dot(n) /(sqrt(m.dot(m))*sqrt(n.dot(n)) ) )
print('{:s}{:8.2f}'.format('dihedral angle = ', psi*180/np.pi ))
dihedral angle = 44.42
This same result can be obtained more easily from geometry; a side view is show in figure 78.
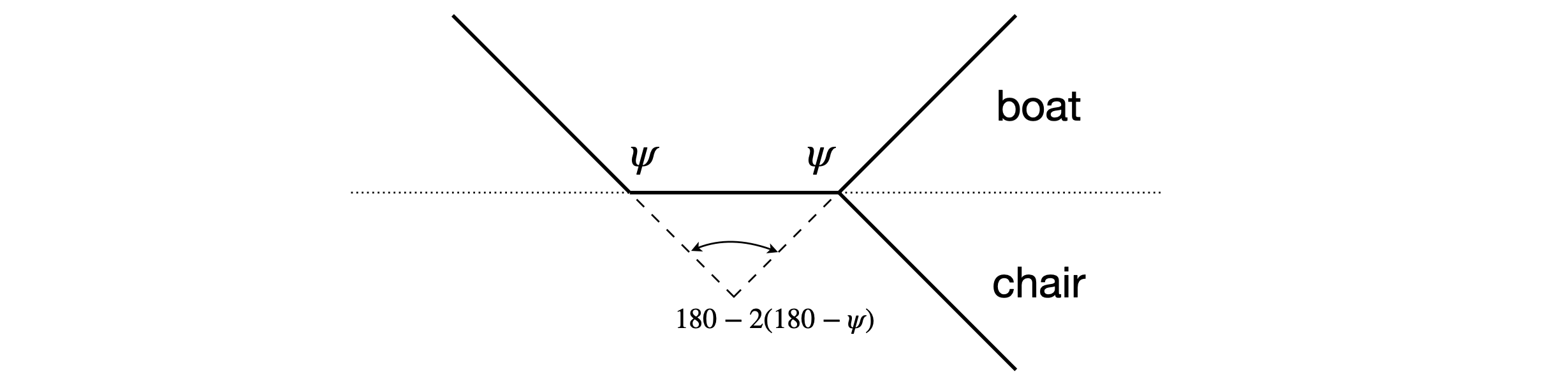
Figure 78. Side view for cyclohexane chair and boat forms.
In this last calculation, if the \(C_4\) atom from the chair form was used the calculated dihedral angle would have been \(180^\text{o}\) because the two planes involved are parallel.
Q55 answer#
The bond N11-C12 lies between the two planes whose torsion (dihedral) angle \(\psi\) is to be calculated. Vectors \(\vec m\) and \(\vec n\) are therefore
so that the vectors point from C8 towards N16. Defining these vectors is the most difficult part as Python can be used to complete the algebra. The script to do this is shown below. A procedure is defined that accepts the atoms as input. Notice that we check that the angle has the correct sign with a function ‘sign’. This function returns \(+1\) or \(-1\) only.
C8 = np.array( [ 120.627 , 4.607 ,38.990 ] )
N11 = np.array( [ 120.292 , 4.951 , 40.228 ] )
C12 = np.array( [ 118.955 , 4.753 , 40.750 ] )
C13 = np.array( [ 117.996 , 5.868 , 40.333 ] )
N16 = np.array( [ 118.093 , 7.008 , 41.010 ] )
#this procedure accepts atom vectors as input
def torsion_angle(a1, a2, a3, a4):
A = a2 - a1
B = a3 - a2
C = a4 - a3
m = np.cross(A, B)
n = np.cross(B, C)
cos_angle = np.dot(m, n)/(np.sqrt( np.dot(n, n) )*np.sqrt( np.dot(m, m) ) )
angle = np.sign(np.dot(n,A))*np.arccos(cos_angle)*180/np.pi
return angle
phi = torsion_angle(C8, N11, C12, C13)
print('{:s}{:8.2f}'.format(' torsion angle =', phi) )
phi=torsion_angle(N11,C12,C13,N16)
print('{:s}{:8.2f}'.format(' torsion angle =', phi) )
torsion angle = -81.90
torsion angle = -78.25
Consulting a Ramachandran plot, such as figure 38, suggests that this small part of the structure belongs to an \(\alpha\) -helix.
Q56 answer#
The Python code to read the pdb is shown below. The order of atoms is always N-CA-C-O followed by the atoms in the residue. These atoms have indices 0,1,2,3 in the lists resA, resB, resC below. The next residue has the same pattern and so on. Three sequential residues are identified and read in in one pass of the data. Residue 35 is needed for part (b). The results are stored and then the torsion angle calculated. There are two chains in this protein so it is necessary to identify which one is to be used. The residue number to is are given in the question.
# Algorithm. Torsion angle
dataname='1TIM.pdb'
numA = 36 # residue 36
numB = numA + 1 # residue B
numC = numA - 1
chain= 'B' # which chain ?
resA = np.zeros((14,3),dtype=float) # as largest residiue trp has 14 atoms, 3 for x,y,z
resB = np.zeros((14,3),dtype=float)
resC = np.zeros((14,3),dtype=float)
with open(dataname) as f:
kA = 0 # counter for index in list resA etc.
kB = 0
kC = 0
for line in f:
new_str=' '.join(line.split())
vals = new_str.split(' ')
if vals[0] == 'ATOM':
if vals[5] == str(numA) and vals[4] == chain:
resA[kA][:] = [vals[6],vals[7],vals[8]]
kA = kA + 1
if vals[5] == str(numB) and vals[4] == chain:
resB[kB][:] = [vals[6],vals[7],vals[8]]
kB = kB + 1
if vals[5] == str(numC) and vals[4] == chain:
resC[kC][:] = [vals[6],vals[7],vals[8]]
kC = kC + 1
f.close()
print('{:s}{:6.3f}'.format('torsion angle = ',torsion_angle(resA[0],resA[1],resA[2],resB[0]) ) )
torsion angle = -14.605
(b) The \(\phi\) angles are those between the N and the next adjacent C\(_\alpha\) atom; the sequence of atoms is C-N-C\(_\alpha\) - C. In figure 48, the first \(\phi\) angle is between N36 and CA36 but the list starts at C35. We use the values already read into the arrays.
print('{:s}{:6.3f}'.format('torsion angle = ',torsion_angle(resC[2],resA[0],resA[1],resA[2]) ) )
torsion angle = -85.957
Q57 answer#
The torsion angle \(v_2\) should be C\(_3'\) endo for A-type DNA, and therefore have an angle from \(0 \to 90^\text{o}\). We can use the torsion_angle(…) code above
C1 = np.array( [ 8.500 , 0.483 , -4.443 ] )
C2 = np.array( [ 8.902 , -0.134, -5.779 ] )
C3 = np.array( [ 8.751 , 1.078 , -6.697 ] )
C4 = np.array( [ 9.378 , 2.172 , -5.835 ] )
C5 = np.array( [ 8.989 , 3.594 , -6.187 ] )
O4 = np.array( [ 8.917 , 1.839 , -4.493 ] )
print('{:s}{:6.3f}'.format('torsion angle = ',torsion_angle(C1,C2,C3,C4) ) )
torsion angle = 42.742
which confirms the A type DNA. Extra confirmation would be that the angle \(\gamma\) is between approximately \(30\) and \(90^\text{o}\).
(b) As a check on the conformation, the C\(_3'\) atom should be above the \(\mathrm{C_4'-O_4'-C_1'}\) plane and on the same side as the C\(_5'\) atom with the C\(_2'\) below it. The plane is defined by the vector \(\vec n\) normal to it. This is the cross product of the \(\vec{O_4-C_5}\) vector and the \(\vec{O_4-C_1}\) vector (see equation 44) and gives the equation of the plane. If \(T\) is at some particular point in the plane, with position vector \(\vec T\) , the distance to a point \(p\) with position vector \(\vec p\), is given by equation 45 which is \(\displaystyle d=\frac{\vec n\cdot(\vec p - \vec T)}{\sqrt{\vec n \cdot \vec n}}\). The Python code is shown below;
n =np.cross( (C4-O4) , (C1-O4) )
T = O4
p = C3
d = np.dot(n,p-T)/np.sqrt( np.dot(n,n) )
print('{:s}{:6.3f}'.format('to C3 d = ',d ) )
d = np.dot(n,C2-T)/np.sqrt(np.dot(n,n) )
print('{:s}{:6.3f}'.format('to C2 d = ',d ) )
d = np.dot(n,C5-T)/np.sqrt(np.dot(n,n) )
print('{:s}{:6.3f}'.format('to C5 d = ',d ) )
to C3 d = 0.495
to C2 d = -0.209
to C5 d = 0.842
Because the distance to C\(_5'\) has the same sign as that to C\(_3'\), these atoms must be on the same side of the sugar ring, making C\(_3'\) endo and C\(_2'\) exo because it has the opposite sign. The configuration has therefore been determined independently, without having to know what the range of torsion angles is supposed to be for a particular configuration.
Q58 answer#
(a) The base shown is guanine which is a purine base. (b) Using the torsion_angle(…) code developed earlier, the calculation becomes very easy. All that is necessary is to put the coordinates into vector form.
O5 = np.array( [ 3.903 , 9.424 , 56.660 ] )
C5 = np.array( [ 3.829 , 10.592 , 57.472 ] )
C4 = np.array( [ 2.610 , 11.430 , 57.099 ] )
C3 = np.array( [ 1.382 , 10.524 , 57.025 ] )
C2 = np.array( [ 1.034 , 10.517 , 55.563 ] )
C1 = np.array( [ 1.598 , 11.813 , 55.022 ] )
O4 = np.array( [ 2.772 , 12.046 , 55.805 ] )
N9 = np.array( [ 1.981 , 11.653 , 53.605 ] )
C4B= np.array( [ 1.493 , 12.380 , 52.547 ] )
print('{:s}{:6.3f}'.format('gamma = ',torsion_angle(O5,C5,C4,C3) ) )
print('{:s}{:6.3f}'.format('v2 = ',torsion_angle(C1,C2,C3,C4) ) )
print('{:s}{:6.3f}'.format('chi = ',torsion_angle(O4,C1,N9,C4B)) )
gamma = 48.314
v2 = -25.033
chi = -124.566
The DNA should be B type because the ring pucker is negative, indicating C\(_2'\) endo configuration. This can be confirmed by calculating the distance above and below the ring as in the previous example. The \(\chi\) torsion angle is large and negative indicating anti configuration.
Exercise: Write a procedure to do a calculation to determine the DNA type given only the coordinate vectors. Calculate the \(\gamma, chi\) and \(v_2\) angles and the absolute configuration, by calculating the distance of the C\(_2'\) and C\(_3'\) atoms above the \(\mathrm{C_4'-O_4'-C_1'}\) plane. Print out a statement of the DNA type as well as C\(_3'\) exo, or endo etc and the angles and distances. Test your algorithm on the PDB entry 1AAY and on other DNA structures in the database.
Q59 answer#
Using the definitions given in the question, the result is
However, the cross product of a vector with itself is zero, hence
Q60 answer#
The expansion of the triple product (see Section 17) gives \(\displaystyle \vec a=\vec\omega \times(\vec\omega \times \vec r)=(\vec\omega\cdot \vec r)\vec\omega-(\vec\omega\cdot \vec\omega)\vec r\)
Since \(\vec r\) and \(\vec\omega\) are perpendicular, their dot product is zero, hence \(\vec a = -(\vec\omega\cdot \vec\omega)\vec r\) and the magnitude of \(a\) is \(|\vec a| = a -\omega^2r = -v^2/r\). As the sign is negative this centripetal acceleration is towards the centre of the orbit.
Q61 answer#
There is no \(k\) component, as motion is only in the \(x-y\) or \(i-j\) plane. The angular velocity, by definition, is perpendicular to this plane, and has a component only in the \(z\) or \(\vec k\) direction where \(\vec\omega = \alpha\vec \omega k\) or \(|\vec \omega |=\alpha\).
The cross product of \(\vec \omega\) and \(\vec r\) is
where in the last step we use the anti-commutative rule \(\boldsymbol k\times \boldsymbol j=-\boldsymbol j\times k =-\boldsymbol i\).
Velocity is the time derivative of position, and the linear velocity at any time \(t\) is
As the cross product, equation A, is the same as the velocity, equation B, we conclude that linear velocity is the cross product of angular velocity and radius vector or \(\vec v=\vec \omega \times \vec r\).


